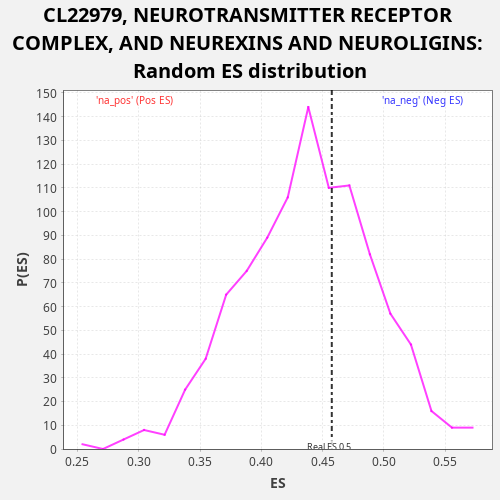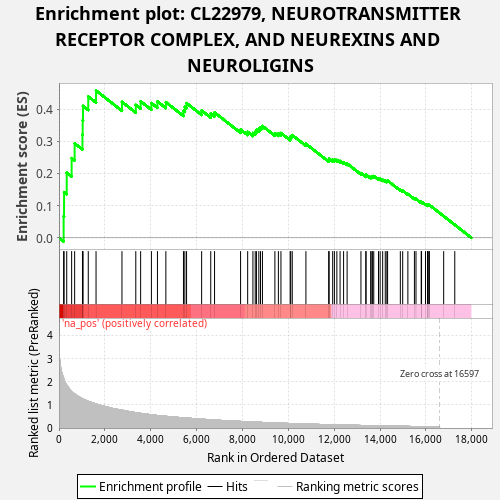
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | COVID-19 positive (controls include untested)__COVID19-both_sexes-C2_v2-04162021 |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.45752683 |
| Normalized Enrichment Score (NES) | 1.0442135 |
| Nominal p-value | 0.361 |
| FDR q-value | 0.74899244 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | HTR3C | 202 | 2.153 | 0.0669 | Yes |
| 2 | NLGN2 | 221 | 2.089 | 0.1418 | Yes |
| 3 | OLFM2 | 337 | 1.844 | 0.2024 | Yes |
| 4 | SLITRK5 | 551 | 1.571 | 0.2475 | Yes |
| 5 | CACNG2 | 682 | 1.464 | 0.2935 | Yes |
| 6 | GRIK4 | 1023 | 1.259 | 0.3202 | Yes |
| 7 | DLG3 | 1033 | 1.253 | 0.3652 | Yes |
| 8 | NETO2 | 1046 | 1.247 | 0.4098 | Yes |
| 9 | GRIN1 | 1276 | 1.152 | 0.4389 | Yes |
| 10 | CACNG5 | 1615 | 1.033 | 0.4575 | Yes |
| 11 | CACNG8 | 2745 | 0.770 | 0.4225 | No |
| 12 | MDGA1 | 3348 | 0.662 | 0.4129 | No |
| 13 | GRIN2A | 3559 | 0.630 | 0.4241 | No |
| 14 | GRID1 | 4029 | 0.570 | 0.4186 | No |
| 15 | GRM5 | 4292 | 0.539 | 0.4235 | No |
| 16 | SYNGAP1 | 4659 | 0.504 | 0.4214 | No |
| 17 | GRM4 | 5427 | 0.438 | 0.3945 | No |
| 18 | CLSTN3 | 5487 | 0.433 | 0.4069 | No |
| 19 | GRIK1 | 5561 | 0.428 | 0.4184 | No |
| 20 | NETO1 | 6219 | 0.381 | 0.3955 | No |
| 21 | GRM8 | 6616 | 0.353 | 0.3862 | No |
| 22 | CHRM3 | 6782 | 0.342 | 0.3895 | No |
| 23 | HOMER2 | 7917 | 0.283 | 0.3364 | No |
| 24 | CNIH3 | 8226 | 0.268 | 0.3290 | No |
| 25 | GRIK3 | 8455 | 0.258 | 0.3256 | No |
| 26 | GRIN2C | 8555 | 0.253 | 0.3293 | No |
| 27 | HTR3A | 8609 | 0.250 | 0.3354 | No |
| 28 | GRM1 | 8712 | 0.246 | 0.3386 | No |
| 29 | NRXN2 | 8786 | 0.243 | 0.3434 | No |
| 30 | CBLN2 | 8870 | 0.239 | 0.3474 | No |
| 31 | SHANK1 | 9413 | 0.217 | 0.3250 | No |
| 32 | GRIK2 | 9565 | 0.210 | 0.3242 | No |
| 33 | FBXO40 | 9677 | 0.206 | 0.3255 | No |
| 34 | DLG4 | 10078 | 0.193 | 0.3102 | No |
| 35 | GRIA4 | 10103 | 0.192 | 0.3158 | No |
| 36 | HTR3D | 10170 | 0.190 | 0.3191 | No |
| 37 | GRIN2D | 10766 | 0.170 | 0.2920 | No |
| 38 | HTR3E | 11758 | 0.141 | 0.2418 | No |
| 39 | SHISA7 | 11778 | 0.141 | 0.2458 | No |
| 40 | NRXN3 | 11933 | 0.137 | 0.2422 | No |
| 41 | VWC2L | 12008 | 0.135 | 0.2430 | No |
| 42 | HTR3B | 12115 | 0.132 | 0.2418 | No |
| 43 | LRRTM1 | 12255 | 0.129 | 0.2388 | No |
| 44 | DLGAP4 | 12408 | 0.126 | 0.2348 | No |
| 45 | NLGN1 | 12562 | 0.122 | 0.2307 | No |
| 46 | DLGAP3 | 13165 | 0.108 | 0.2010 | No |
| 47 | GRIA1 | 13378 | 0.104 | 0.1930 | No |
| 48 | HOMER1 | 13387 | 0.103 | 0.1963 | No |
| 49 | GRIA2 | 13587 | 0.099 | 0.1888 | No |
| 50 | GRIN3A | 13640 | 0.098 | 0.1894 | No |
| 51 | GRIN2B | 13674 | 0.097 | 0.1911 | No |
| 52 | SHANK3 | 13724 | 0.096 | 0.1919 | No |
| 53 | SHANK2 | 13931 | 0.092 | 0.1837 | No |
| 54 | TJAP1 | 14002 | 0.091 | 0.1831 | No |
| 55 | NLGN4X | 14112 | 0.088 | 0.1802 | No |
| 56 | GRM7 | 14225 | 0.086 | 0.1771 | No |
| 57 | NRXN1 | 14295 | 0.085 | 0.1763 | No |
| 58 | DLGAP1 | 14327 | 0.084 | 0.1776 | No |
| 59 | NLGN3 | 14882 | 0.074 | 0.1494 | No |
| 60 | HOMER3 | 14985 | 0.072 | 0.1463 | No |
| 61 | CBLN4 | 15211 | 0.068 | 0.1362 | No |
| 62 | CHRM5 | 15496 | 0.063 | 0.1226 | No |
| 63 | SHISA9 | 15565 | 0.061 | 0.1211 | No |
| 64 | GRID2 | 15793 | 0.057 | 0.1104 | No |
| 65 | LHFPL4 | 15807 | 0.056 | 0.1118 | No |
| 66 | CNIH2 | 15979 | 0.053 | 0.1041 | No |
| 67 | GRIN3B | 16067 | 0.051 | 0.1011 | No |
| 68 | CDH9 | 16074 | 0.051 | 0.1026 | No |
| 69 | PTCHD1 | 16101 | 0.050 | 0.1030 | No |
| 70 | CNTN4 | 16151 | 0.049 | 0.1020 | No |
| 71 | CBLN1 | 16772 | -0.000 | 0.0674 | No |
| 72 | DLG2 | 17256 | -0.000 | 0.0404 | No |